De novo Protein Design | Ancestral Protein Design | Consensus Design | Computational Protein Design
X-ray crystallography | Structural Modelling | Molecular Dynamics Simulations
ITC | SPR | DSF
De novo Protein Design and Protein Optimisation
Protein hallucination & diffusion
We are using modern protein backbone generation tools (including RFDiffusion, RFDesign & AFDesign) for the generation of new-to-nature proteins. This include protein folds never observed in nature, and de novo binder proteins to a range of medically- and biotechnologically-relevant proteins.
Protein optimisation & stabilisation
We apply a range of tools for the generation of stablised protein variants that express well in common expression hosts while retaining target functions. We focus on valuable proteins for biotechnology, medicine and synthetic biology.
Protein Evolution
Similar to the process of evolution at the level of organisms, proteins gain new functions and characteristics through gene-duplication events, the accumulation of mutations (genetic diversification) and natural selection. Understanding the fundamental mechanisms through which proteins gain new functions can provide insight into the factors that drive functional diversification of protein families, but also can guide protein engineering efforts by highlighting the factors that drive or hinder the emergence and improvement of new functions and/or properties.
The evolution of an enzyme from a non-catalytic binding protein
I have had the pleasure of working on a project studying the natural evolution of an enzyme, cyclohexadienyl dehydratase, from an ancestral protein that was non-catalytic. Earlier evolutionary biochemistry studies had highlighted the transition of one enzyme to another enzymatic activity using directed evolution and ancestral reconstruction approaches, but no one had really studied the mechanisms underlying the emergence of new enzyme activity in proteins folds that are non-catalytic.
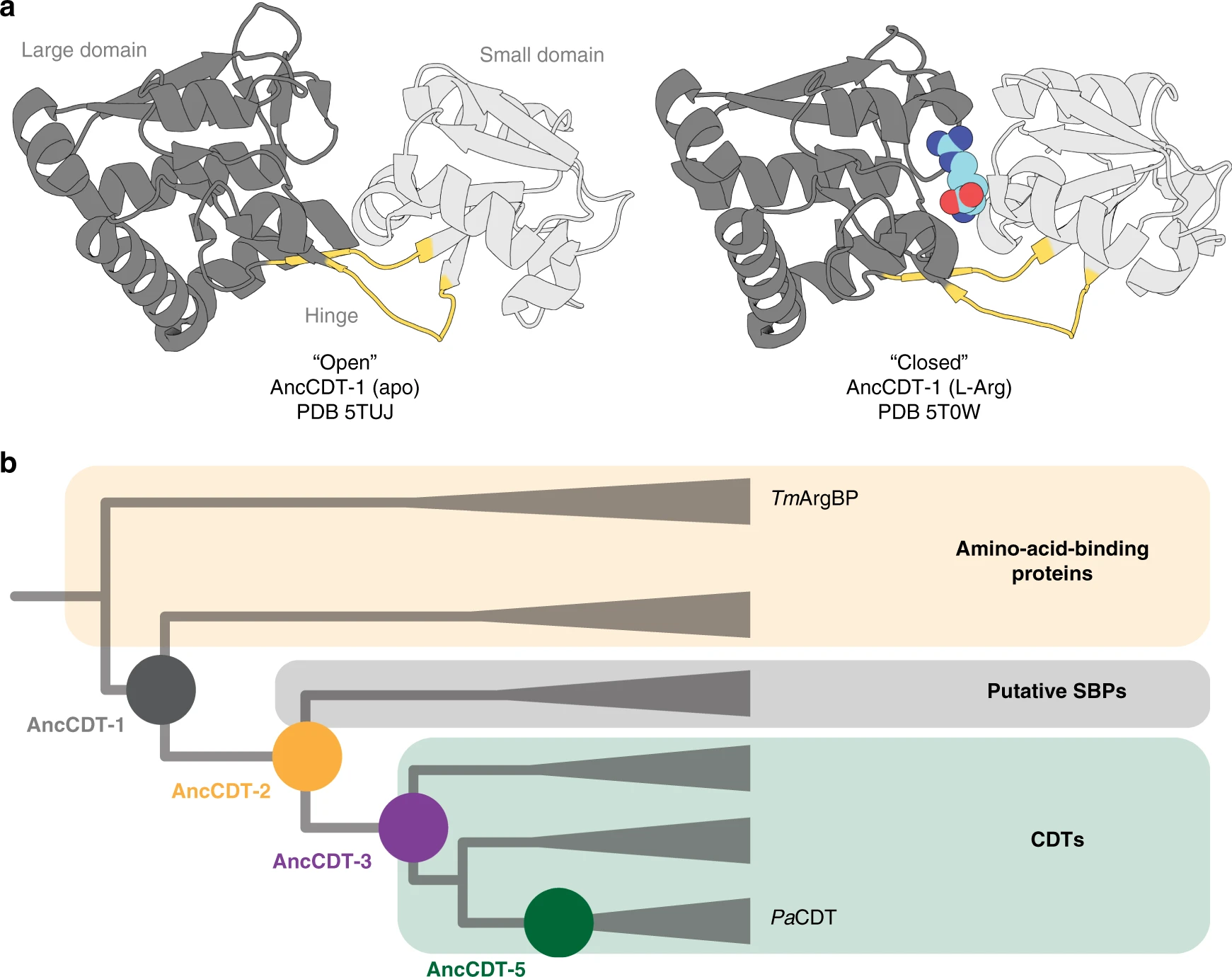
In a study led by Ben Clifton, we showed that the enzyme cyclohexadienyl dehydratase evolved from an ancestral solute-binding protein, and that the new enzyme activity emerged and improved through distinct steps including (i) the introduction of key catalytic residues into the binding site of the ancestral protein, (ii) the reshaping of the active site cavity and development of hydrogen bonding networks around the core of the protein, and (iii) the accumulation of remote mutations that likely altered the conformational sampling of the enzyme (favouring the more closed, catalytically-relevant state).
In a follow up study, we collaborated with Gottfried Otting (ANU) and Daniella Goldfarb (Weizman Institute of Science, Isreal), to explore how the sampling of open and closed states changed during the evolution from a non-catlytic binding protein to an enzyme. By comparing enzyme kinetic data with molecular dynamics simulations and double electron-electron resonance spectroscopy measurements (on tagged variants of the proteins), we showed that: (i) the ancestral solute-binding protein sampled both open and closed states, typical of proteins with the periplasmic binding protein fold; (ii) the intermediate ancestors sampled mainly wide open states which were likely unproductive to catalysis and (iii) the modern enzyme sampled more compact states, which likely contributed to the observed increase in catalytic efficiency.
In the third paper in this series, we investigated the role that oligomerisation and a C-terminal extension had in the optimisation CDT activity (modern CDT is a trimer, while AncCDT-5 and other proteins are monomers). We introduced subtitutions found in the CDT trimer-forming interface, and reintroduced these into AncCDT-5. As we introduced these substitutinos, we observed the transition from monomer to trimer. The initial formation of the trimeric state was associated with a decrease in the affinity of the enzyme for the substrate. Finally, we showed that removal of a C-terminal extension found in modern CDT yielded a protein that had a catalytic turn-over similar to AncCDT-5, suggesting that this region played a role in further tuning catalysis. Molecular dynamics simulations showed how the formation of the trimer, and the C-terminal extension, restricted open-closed motions in the protein to minimise sampling of unproductive open states.
Protein Dynamics
My life changed the moment that I started seeing proteins as dynamic species, rather than the static blobs that were portrayed in my high-school text books. Recognising the important role that dynamics has in determining the properties of proteins (including thermostability, activity, interactions with other proteins, involvement in signalling pathways etc) is vital when studying and engineering proteins - and makes studying these molecules even more fascinating.
As described above, our work on cyclohyexadienyl dehydratase showed how the sampling of different conformational states has been important during the evolution of a new enzyme activity. Many of the other projects that I have been involved in have had a particular focus on studying how properties of a protein are influenced by the way a protein moves.
In my first publication, I worked under the supervision of Ben Corry (ANU) to study the dynamics of the fenestrations of voltage-gated sodium channels. These fenestrations (channels that lead from the membrane to the central cavity of the protein) are important for allowing the passage of small molecule drugs that are used to block sodium channels. Our molecular dynamics simulation study highlighted the dynamic nature of these fenestrations, and highlighted key residues that lined the tunnels which likely control access to the central cavity of the protein. Interestingly, we saw that lipids from the membrane often protruded into these fenestrations.
Structures and dynamics of non-ribosomal peptide synthesis proteins
Structural Immunology
Vibrio natriegens as an alternate expression host
We are currently leveraging the rapid doubling time and high protein production capabilities of Vibrio natriegens cells as an alternate protein expression host. In addition, we are exploring ways to genetically manipulate Vibrio natriegens to enhance the production of valuable compounds and proteins.
Other Protein Engineering Projects
Protein-based Sensor Design
- Kaczmarski JA, Mitchell JA, Spence MA, Vongsouthi V, Jackson CJ. 2019. Structural and evolutionary approaches to the design and optimization of fluorescence-based small molecule biosensors. Curr Opin Struct Biol 57:31–38.
Cyanobacterial carbon-concentrating mechanisms
- Kaczmarski JA, Hong NS, Mukherjee B, Wey LT, Rourke L, Förster B, Peat TS, Price GD, Jackson CJ. 2019. Structural basis for the allosteric regulation of the SbtA bicarbonate transporter by the PII-like protein, SbtB, from Cyanobium sp. PCC7001. Biochemistry 58:5030–5039.